Authors
Johanne U. Hermansen, Weikaixin Kong, Andrea M. Brodersen, Yanping Yin, Aleksandra Urban, Idun D. Rein, Liye He, Rudi Agius, Rebecca Svanberg Teglgaard, Juho Rousu, Christian Brieghel, Sabina Kersting, Mark-David Levin, Hoa T. T. Tran, Mattias Mattsson, Juha Ranti, Gerrit-Jan Veldhuis, Caspar da Cunha-Bang, Rogier Mous, Julie Dubois, Arnon P. Kater, Jorrit M. Enserink, Carsten U. Niemann, Tero Aittokallio, Sigrid S. Skånland.
Background:
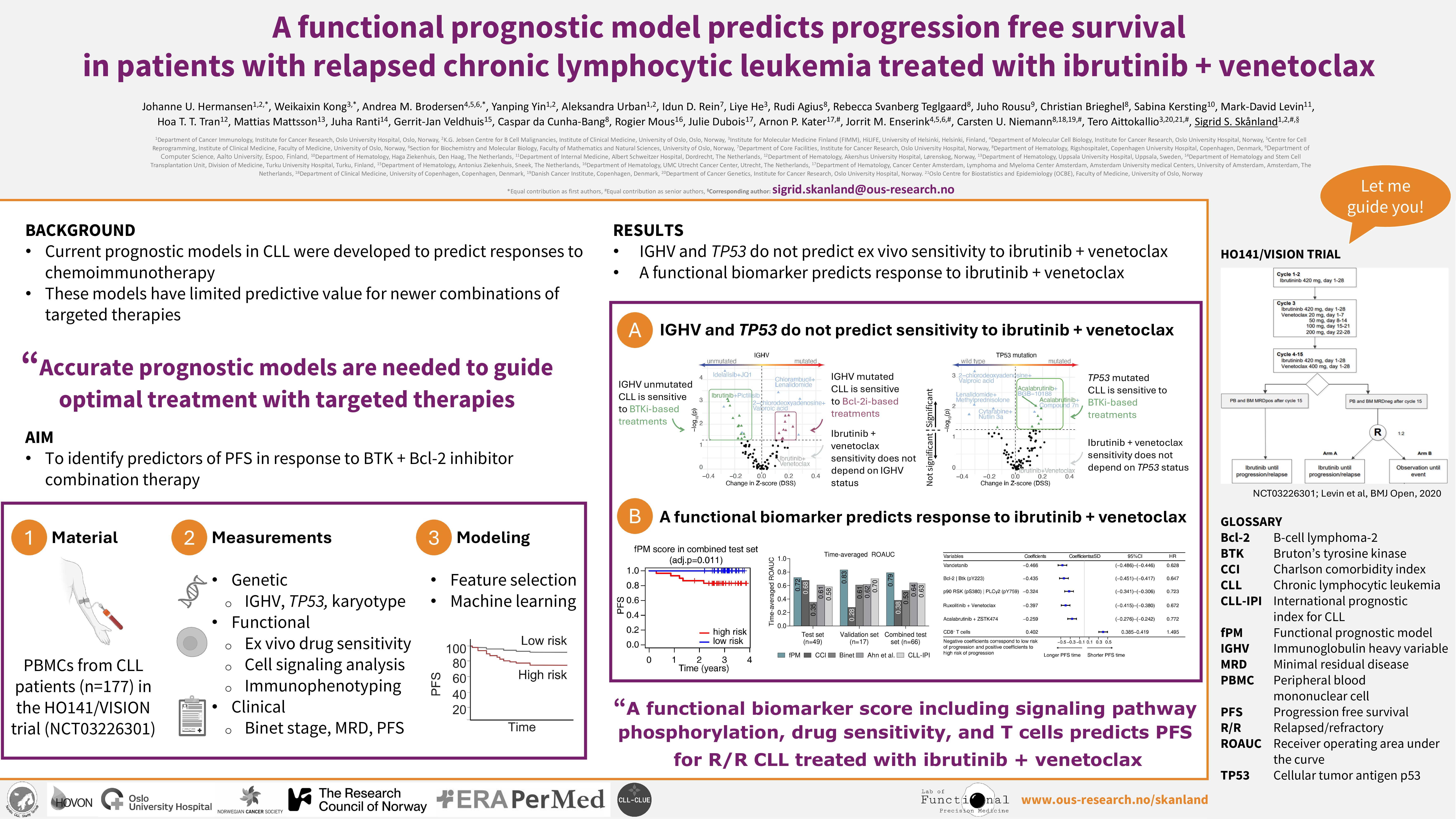
Targeted therapies have made a breakthrough in treatment of chronic lymphocytic leukemia (CLL), but many patients develop resistance, experience side effects, or relapse during therapy. Since traditional biomarkers for chemoimmunotherapy have failed to predict clinical outcomes on time-limited combination targeted therapies, new clinicobiological features are warranted. HO141/VISION (NCT03226301) is a randomized phase II trial that evaluates the efficacy of minimal residual disease (MRD) guided combination treatment with the Bcl-2 inhibitor (Bcl-2i) venetoclax and the BTK inhibitor (BTKi) ibrutinib in relapsed/refractory (R/R) patients with CLL.
Aims:
i) Identify associations between functional, genetic, and clinical characteristics in CLL.
ii) Define biomarkers that predict progression free survival (PFS) on the HO141/VISION trial.
Methods:
Peripheral blood mononuclear cells (PBMCs) were collected from patients with CLL (n = 177) enrolled in the HO141/VISION trial prior to start of treatment. The samples were subjected to functional analyses: immunophenotyping (6 surface markers for B-, T- and NK-cell subpopulations) and (phospho)protein profiling (31 markers) by flow cytometry, and drug sensitivity screening (95 single agents, 87 combinations) using the CellTiter-Glo luminescent cell viability assay. Association analyses between functional, genetic (IGHV mutational status, TP53 aberrations, karyotypes), and clinical (Binet stage, MRD, PFS) features were investigated using the Wilcoxon rank sum test. SHAP values were calculated from random forest to understand the directionality and importance of each feature in predicting ex vivo drug sensitivity. To identify a biomarker predictive of PFS (20 events), the patient samples were divided into a training set (n=80) and a test set (n=77) stratified on IGHV and TP53 mutational status, as well as an independent validation set (n=20). The (phospho)proteins detected by flow cytometry were paired and used as binary features. Univariate Cox regression and permutation testing in random survival forest identified the most predictive features using 10-cross validation in the training set (p < 0.1), and the overlapping features from the two selection methods were utilized as a multi-modal signature to predict PFS with various machine learning models.
Results:
Genetic subgroups of CLL showed distinct ex vivo drug sensitivities. We confirmed that TP53 aberrant CLL cells are less sensitive to chemotherapy and nutlin 3a relative to TP53 wild-type CLL cells. We identified a trend towards higher sensitivity to BTKi and PI3Ki single agents or combinations in CLL cells from high-risk (TP53 aberrant, IGHV unmutated) patients, and to Bcl-2i/combinations in CLL cells from low-risk (IGHV mutated) patients. We found that phosphorylation of PLC2 (pY759), which is downstream of BTK, was significantly higher in CLL cells from low-risk patients than in CLL cells from high-risk patients. This may explain the reduced ex vivo sensitivity to BTKi in this patient group. Association studies of functional, genetic, and clinical features showed that (phospho)protein profiles had the highest predictive power on ex vivo drug sensitivity.
We used both functional and genetic features to predict patient-specific PFS. Our machine learning model selected only functional features as most predictive of PFS, ex vivo sensitivity to vandetanib (VEGFR/EGFR inhibitor), ruxolitinib (JAK1/2 inhibitor) + venetoclax (Bcl-2i), and acalabrutinib (BTKi) + ZSTK474 (PI3Ki); relative expression/phosphorylation level of Bcl-2|BTK (pY223) and p90RSK (pS380)|PLCγ2 (pY759); and a high proportion of CD8+ T cells. These findings demonstrate that patterns of ex vivo drug sensitivity profiles can predict response to a given but different treatment. The resulting functional prognostic model (fPM) demonstrated increased and robust performance compared to the CLL international prognostic index (CLL-IPI) and other reported scores.
Summary/conclusion:
i) Genetic subgroups of CLL show distinct ex vivo drug sensitivities, and protein profiles have the highest predictive power on ex vivo drug sensitivity
ii) Functional features predict PFS in patients with R/R CLL treated with ibrutinib + venetoclax, warranting clinical validation also for frontline treatment of CLL.
Keywords : Biomarker, targeted therapy, outcome prediction
Please indicate how this research was funded. :
Please indicate the name of the funding organization.: