Authors
Kamila Stránská, Jana Štrignerová, Kristýna Taušová, Jana Bruknerová, Hana Skuhrová Francová, Jakub Paweł Porc, Tomáš Reigl, Veronika Navrkalová, Yvona Brychtová, Anna Panovská, Michael Doubek, Šárka Pospíšilová, Karla Plevová.
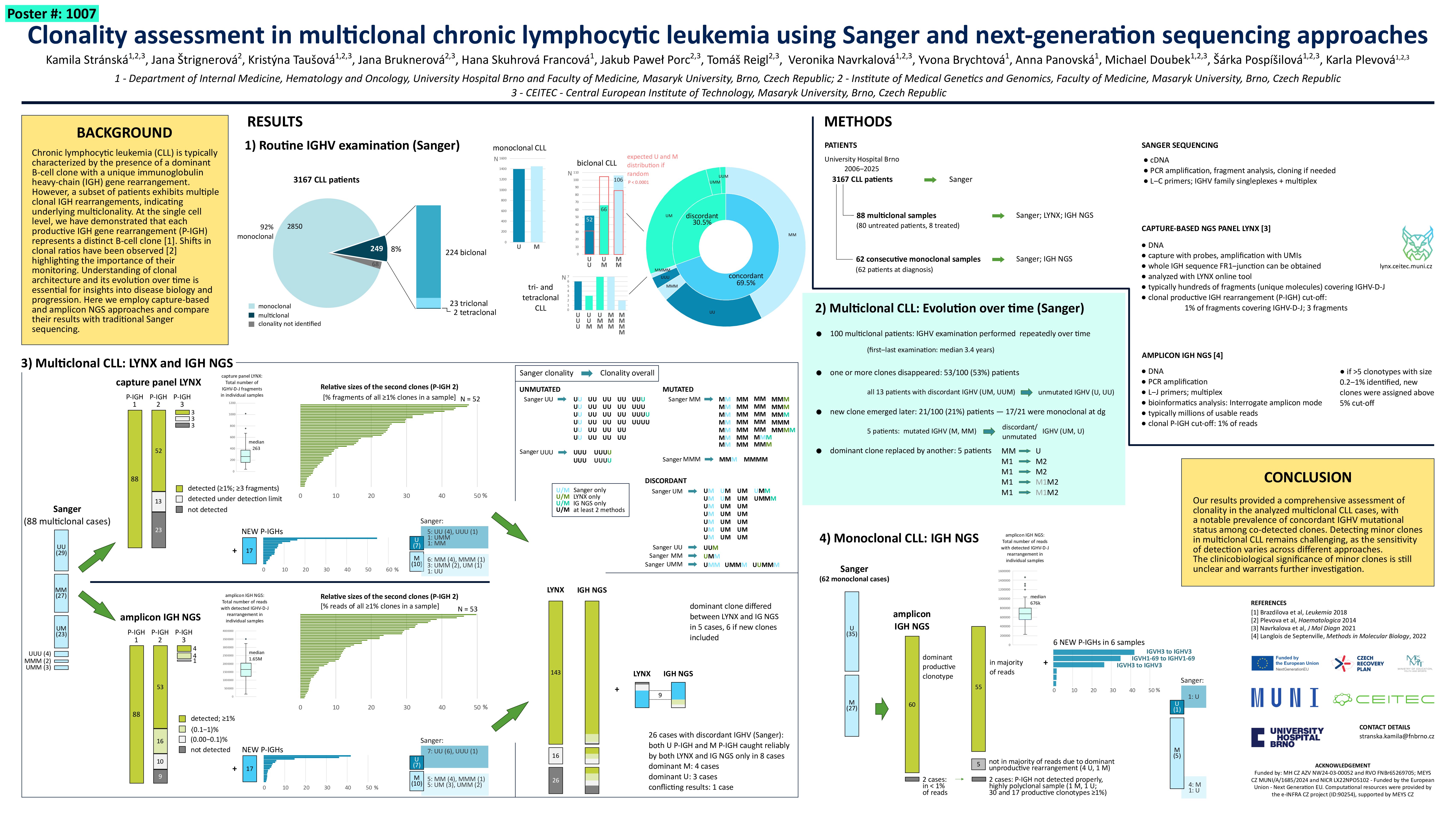
Chronic lymphocytic leukemia (CLL) is typically characterized by the presence of a dominant B-cell clone with a unique immunoglobulin heavy-chain (IGH) gene rearrangement. However, a subset of patients exhibits multiple clonal IGH rearrangements, reflecting underlying multiclonality. Understanding the clonal architecture and its evolution over time is essential for insights into disease biology and progression.
In a cohort of 3,167 CLL patients, routine diagnostic assessment of IGHV mutational status was performed using cDNA and Sanger sequencing. Among these, 249 patients (8%) exhibited multiple clonal productive IGH gene rearrangements. Specifically, we identified 224 biclonal, 23 triclonal, and 2 tetraclonal CLL cases. In 173/249 cases, the co-detected clones were concordantly mutated (115) or unmutated (58). The remaining 76 cases showed discordant mutational status among the clones. IGHV mutational status was examined repeatedly over time in 100/249 multiclonal CLL patients, with a median interval of 3.4 years between the first and last examinations. Over time, one or more clones disappeared in 53 patients, while in 21 patients a new clone emerged later (17/21 patients were monoclonal at diagnosis). In 5 cases, the original dominant clone was replaced by another.
To get deeper insight into the genomic clonal composition of multiclonal CLL cases, we analyzed tumor DNA from a representative cohort of 87 such cases (35% of the multiclonal cohort) with capture-based NGS panel LYNX. This panel allows for detection of markers associated with the most common lymphoid malignancies, including antigen receptor rearrangements. In parallel, we compared detected clonotypes with results from amplicon IGH NGS. The median number of fragments covering complete IGH rearrangements in the LYNX panel was 248, while the median number of usable reads in IGH NGS was 1.7 million. The most abundant clonotype expected was detected in all 87 cases using both methods. Using LYNX, a second expected clone was confidently detected in 53 out of 87 (61%) cases, and in an additional 10 cases below the confident detection threshold (defined as ≥3 fragments and >1% of clonal productive IGH rearrangements). IGH NGS identified the expected second clone in 53/87 (61%) cases at ≥1% of productive IGH rearrangements, and in other 22 cases at levels below 1%. Using either method, we identified additional clonal rearrangements (>1%) that were missed by Sanger sequencing—11 cases using LYNX and 17 using IGH NGS. Notably, in cases where the Sanger-identified clones had concordant IGHV mutational status, all newly identified rearrangements also matched in mutational status: 11 in LYNX and 12 in IGH NGS.
We also performed amplicon IGH NGS analysis on a cohort of 62 consecutive CLL cases seemingly monoclonal by Sanger IGH analysis. In 60 out of 62 cases, the expected rearrangement was the dominant clone. The remaining two cases exhibited a highly polyclonal pattern. In 6 of the 60 cases, an additional clone representing more than 1% of productive IGH rearrangements was detected. In 5 of these 6 cases, the newly identified clone had a concordant IGHV mutational status with the concurrent dominant clone. Notably, three of the additional clonal rearrangements had a relative size >25%, and all three belonged to the same IGHV family as the respective dominant rearangements.
Our results provided a coherent clonality assessment in the analyzed multiclonal CLL cases, with notable prevalence of concordant IGHV mutational status among co-detected clones.
Funded by: MH CZ AZV NW24-03-00052 and RVO FNBr65269705; MEYS CZ MUNI/A/1685/2024 and NICR LX22NPO5102 – Funded by the European Union – Next Generation EU. Computational resources were provided by the e-INFRA CZ project (ID:90254), supported by MEYS CZ.
Keywords : clonality, IGH rearrangement, NGS
Please indicate how this research was funded. : Funded by: MH CZ AZV NW24-03-00052 and RVO FNBr65269705; MEYS CZ MUNI/A/1685/2024 and NICR LX22NPO5102 – Funded by the European Union – Next Generation EU. Computational resources were provided by the e-INFRA CZ project (ID:90254), supported by MEYS CZ.
Please indicate the name of the funding organization.: The Ministry of Health Czech Republic: AZV NW24-03-00052
The Ministry of Health Czech Republic: RVO FNBr65269705
The Ministry of Education, Youth and Sports, Czech Republic: MUNI/A/1685/2024
The Ministry of Education, Youth and Sports, Czech Republic: NICR LX22NPO5102 – Funded by the European Union – Next Generation EU
The Ministry of Education, Youth and Sports, Czech Republic: e-INFRA CZ project (ID:90254)