Authors
Mohammad Al-Agil, Samora Hunter, Konstantina Dimitrakopoulou, David Brawand, Styliani Bouziana, Emil Kumar, Jin-Sup Shin, Suzanne Arulogun, James Teo, Anwar Alhaq, Piers EM Patten.
Rationale
Infection (including bacterial, viral and opportunistic infection) remains a major cause of morbidity and mortality in patients with chronic lymphocytic leukaemia (CLL). Events such as the COVID-19 pandemic further underscored the vulnerability of this population and the need for improved infection risk stratification. However, quantifying individual infection risk remains challenging due to the fragmented nature of clinical data and the absence of predictive tools that integrate real world complexity.
Objectives
To develop a clinically applicable machine learning model that predicts 2-year infection risk in patients with CLL, using routinely collected structured and unstructured electronic patient records (EPRs). The goal was to support early identification of high-risk individuals and enable proactive infection prevention strategies in routine CLL care.
Methods
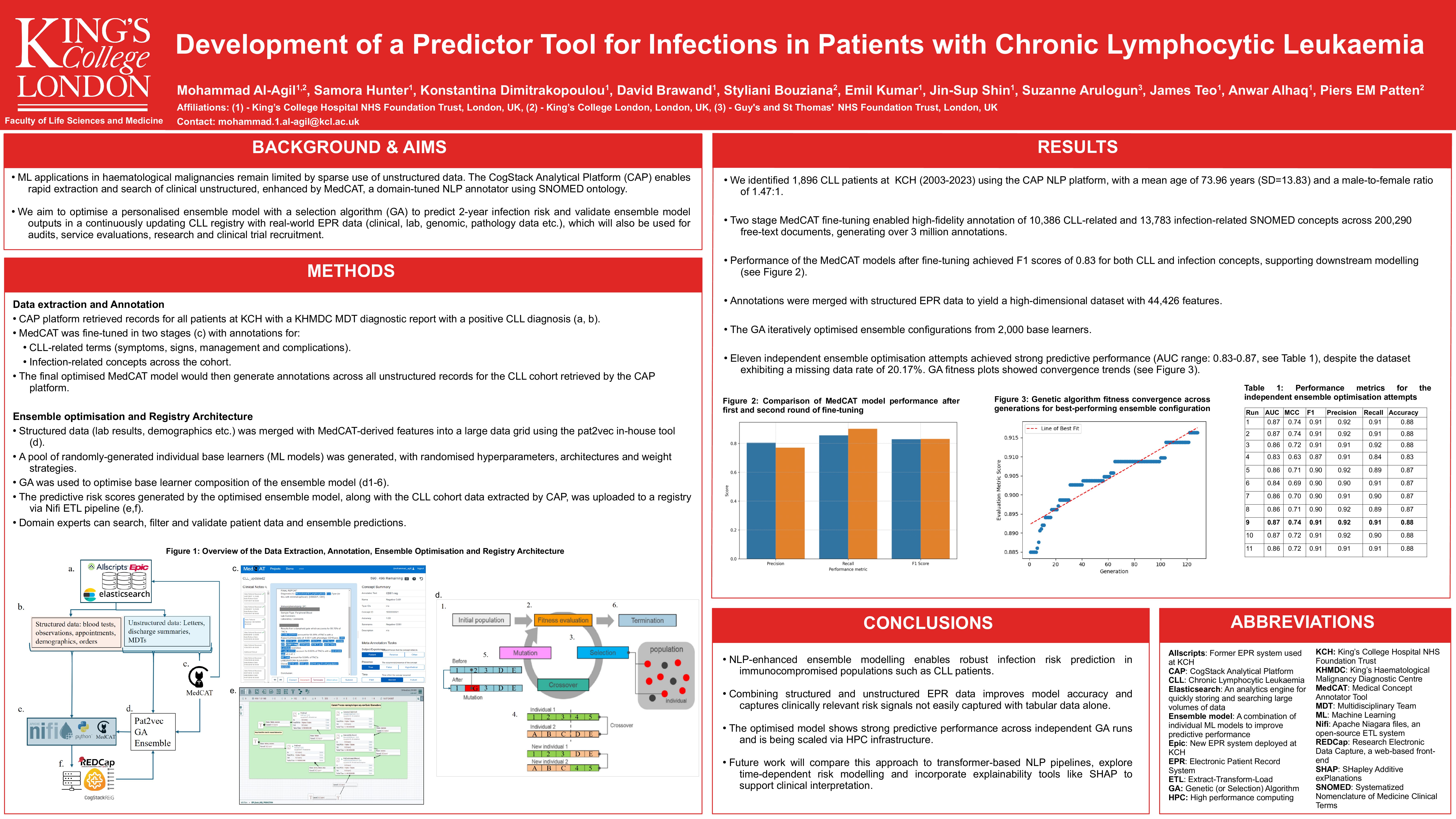
Using the CogStack natural language processing (NLP) platform, 1,896 CLL patients (2003 – 2023) were identified at King’s College Hospital. MedCAT was fine-tuned in two stages: (1) to detect CLL-related concepts (10,386 SNOMED terms) and (2) to identify infections (13,783 terms), producing high-fidelity annotations from 200,290 unstructured documents. Structured variables and NLP-derived features were processed via the pat2vec module. A selection algorithm (SA) optimised ensemble configurations from 2,000 base learners including tree-based, boosting, and neural models to predict 2-year infection risk, using varied hyperparameters and ensemble weighting strategies.
Results
MedCAT yielded F1 scores of 0.829 (for CLL-related concepts) and 0.831 (for infection-related terms), enabling over 3 million high-fidelity annotations from unstructured clinical text. The final ensemble model, trained to predict 2-year infection risk in CLL patients, demonstrated strong predictive performance (AUC: 0.859, F1: 0.904, Accuracy: 0.872), indicating a high ability to correctly identify patients likely to develop infections. Three independent optimisation runs confirmed the model’s robustness (AUC range: 0.793–0.859). Integrating NLP-derived features significantly improved predictive accuracy compared to structured data alone, highlighting the value of leveraging free-text EPR information in forecasting infection risk.
Conclusion
Scalable tools for identifying CLL patients at heightened risk of infection are essential for enabling timely, preventive interventions. This study demonstrates the feasibility of combining structured and unstructured EPR data to build robust, clinically relevant risk models. The SA-optimised ensemble enriched with domain-specific NLP features, offers a data-driven solution for use in routine care and shows promise for broader application across immunocompromised populations. Future work will benchmark this approach against transformer-based NLP systems and evaluate time-dependent modelling strategies.
Keywords : Machine learning, Natural language processing, Infection risk prediction
Please indicate how this research was funded. : A mixture of pharmaceutical funding and charity funding.
Please indicate the name of the funding organization.: