Authors
Pedro Faria Zeni, M. Medkova, L.Janska, J.Pospisil, E.Hoferkova, D.Filip, S.Sharma, L. Ondrisova, W.K.Jesionek, N.Blavet, M.Boudny, N.M.Varadarajan, V.Seda Obrdlik, S.Vanacova, J.Lu, T.Zenz, G.Abrantes Diaz, S.Vinga, M.Hortova-Kohoutkova, K.Kupcova, A.Maiques-Diaz, M. Doubek, V.Vasileiou, F.Psomopoulos, P.Nick, O.Havranek, Jan Fric, J.Martin-Subero, J.Balounova, R.Sedlacek, M.Mraz.
Background
Short non-coding RNAs, namely microRNAs, are well known to influence CLL biology, but the role of long non-coding RNAs (lncRNAs) remains unclear. This is largely due to lncRNAs’ complex mechanisms of action depending on interaction partners (DNA/RNA/proteins).
Results
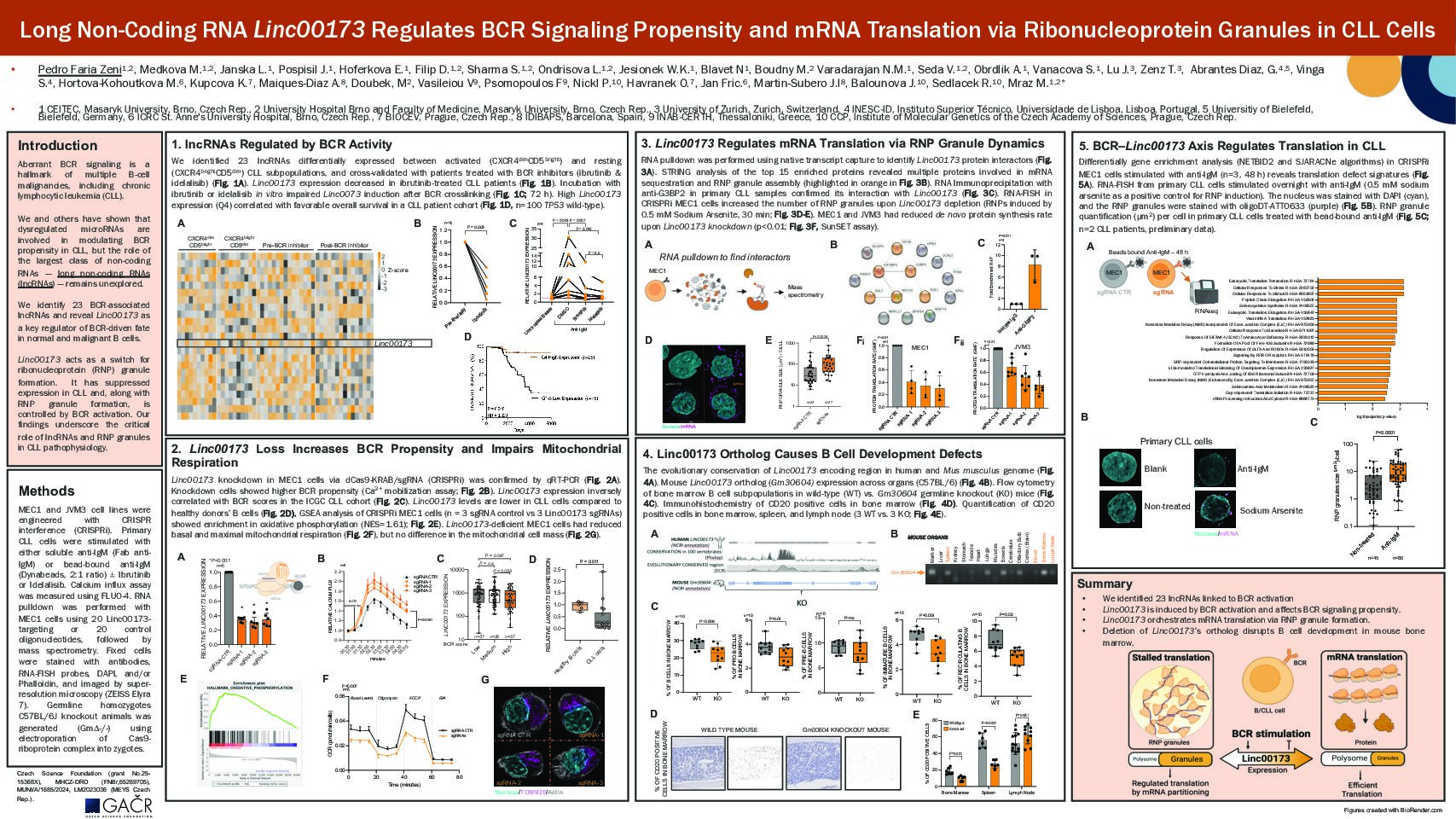
To screen for lncRNAs involved in microenvironmental interactions, we identified lncRNAs differentially expressed in sorted intraclonal CLL subpopulations recently activated in immune niches vs. resting cells (CXCR4dimCD5bright vs. CXCR4brightCD5dim, n=10 pairs) and in CLL cells obtained before and during ~1-3 months of BCR-inhibitor treatment (ibrutinib n=5; idelalisib n=10). Using this strategy, we identified 7 intergenic lncRNAs with changed expression after CLL activation and BCR inhibition. Based on manual inspection of gene annotation, intergenic LINC00173 was selected for further studies, also considering its association with prognosis of multiple tumors (May-Hau et al.,2022). LINC00173 was induced (6.7-fold) in CLL samples by BCR activation in vitro (bead-bound anti-IgM, n=9,72hrs; P=0.003), and this was blocked by ibrutinib or idelalisib treatment. To test its function, we generated MEC1 cells with LINC00173 knockdown (KD) using CRISPR-interference (3 sgRNAs vs. control sgRNA). We hypothesized that LINC00173 might regulate BCR signal propensity and/or CLL activation. Indeed, LINC00173 KD enhanced the MEC1 cells’ responsiveness to BCR crosslinking (anti-IgM, Ca2+ flux assay, n=5, P< 0.001), and the magnitude was comparable to manipulating proteins forming BCR signalosome (SYK, GAB1). The analysis of CLL transcriptomes (ICGC-CLL) revealed significantly higher BCR activity (gene signatures score) in CLL samples with lower vs. higher LINC00173 levels (n=57 vs. 57,P=0.04). Additionally, CLL patients with lower LINC00173 expression had shorter OS in our CLL cohort (n=100 [chemoimmunotherapy era]; P=0.04, median OS: 9.7 vs. 16.8 years), and in ICGC-CLL dataset (n=172). Interestingly, CLL cells (n=18) exhibit ~3-fold lower LINC00173 levels than peripheral blood B cells from healthy donors (n=5; P=0.01). Altogether, this demonstrates that LINC00173 is induced by BCR and also limits BCR signaling propensity. To further dissect the LINC00173 functions, we conducted mRNA profiling in the MEC1 KD/control cells, revealing differential expression of 470 mRNAs (n=3, fold-change≥1.5; Padj < 0.05). This included repression of “negative regulation of lymphocyte activation” in KD cells (NES=-0.42;Padj=0.01), such as PTEN and PTPN22 phosphatases. Moreover, this also revealed the upregulation of 87 mitochondrial genes (NES=2.6;Padj=1.5e⁻10). However, in contrast to transcriptomic data, MEC1 KD cells had reduced (P < 0.05) basal mitochondrial respiration (-25%) and maximal respiration capacity (-40%; n=6, Seahorse assay). These results suggest a compensatory transcriptional response to a functionally deficient metabolic capacity. To understand this, we performed an RNA pulldown (20 probes binding LINC00173) followed by mass spectrometry, which revealed 23 proteins interacting with LINC00173 (n=3, P< 0.05). Notably, this included 5 proteins known to interact and form ribonucleoprotein (RNP) granules involved in mRNA translation control (G3BP2, MOV10, RALY, IGF2BP3, EIF4E), but no proteins annotated as BCR regulators. In line with this, MEC1 KD cells had a higher amount of RNP granules (~3-fold,P < 0.001) when exposed to stress conditions (0.5mM sodium arsenite) and ~65% reduction in de novo protein synthesis in comparison to control cells (SunSET assay; n=4,P < 0.001). Collectively, this suggests that LINC00173 induction by BCR allows for increased protein synthesis via modulating RNPs formation. Next, we investigated LINC00173’s impact on normal B cell fate. We have generated a germline knockout (KO) of mouse LINC00173 ortholog (Gm30604). Gm30604-/- homozygous mice are viable, fertile, and macroscopically normal. The 12-week-old Gm30604-/- mice contained ~20% less B cell numbers in the bone marrow (-24% proB, -13% PreB, -32% immature, -31% recirculating B cells) and 9% less splenic B cells compared to wild-type mice (P < 0.01, n=10 KO vs. 10 WT). Intriguingly, Gm30604-/- mice had an expansion of peritoneal B1b cells (42% increase,P=0.01). This is in line with increased BCR signaling in B1 cells and decreased metabolic/protein synthesis capacity or elimination of BCR-overreactive bone marrow B cells. The KO mice phenotype is under investigation, including the evaluation of any B cell malignancy development with aging.
Conclusion
We have identified the first lncRNA that regulates BCR signaling propensity and impacts protein production capacity after CLL activation. We generated a novel knockout mouse model revealing the impact of LINC00173 on normal B cell production in the bone marrow and B1 cell expansion in the peritoneum.
Supported by: Czech Science Foundation (grant No.25-15368X), MHCZ-DRO(FNBr,65269705), MUNI/A/1685/2024, LM2023036 (MEYS Czech Rep.).
Keywords : BCR, lncRNA, RNP-granules
Please indicate how this research was funded. : Supported by: Czech Science Foundation (grant No.25-15368X), MH CZ-DRO (FNBr,65269705), MUNI/A/1685/2024, LM2023036 (MEYS Czech Rep.).
Please indicate the name of the funding organization.: Czech Science Foundation, Ministry of Health of the Czech Rep, Masaryk University, MEYS